Credit: Laws of Nature (2024). DOI: 10.1038/s41596-024-01046-3
A research team led by scientists at the University of California, Riverside, has developed a computational methodology for analyzing large data sets in the field of metabolomics, the study of small molecules found in of cells, biofluids, cells and the whole environment.
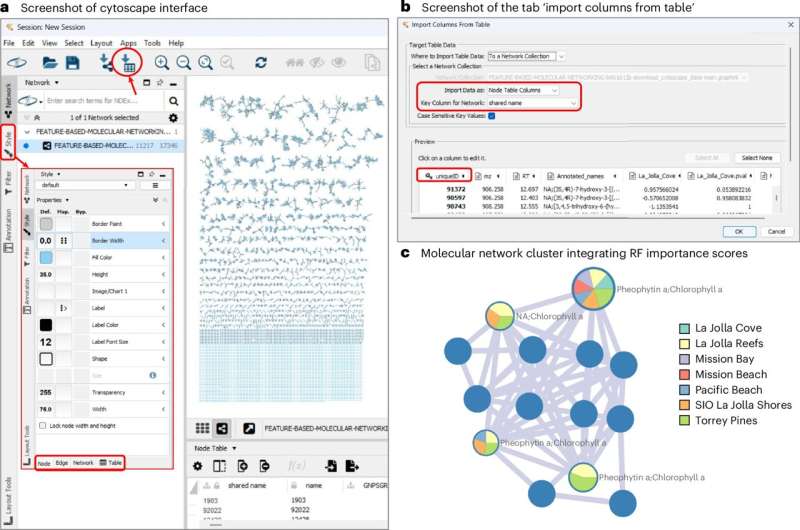
Recently, the team used this new computer tool to analyze pollutants in ocean waters in Southern California. The team quickly took chemical profiles of the coastal environment and highlighted potential sources of pollution.
“We are interested in understanding how such waste is introduced into the environment,” said Daniel Petras, assistant professor of biochemistry at UC Riverside, who led the research team. “Determining which molecules in the ocean are important for the health of the ecosystem is not easy because of the chemical diversity of the ocean. The protocol we developed speeds up the process This much. Sorting data effectively means we can understand the problems associated with ocean pollution.”
Petras and his colleagues report in the journal Laws of Nature that their protocol is designed not only for experienced researchers but also for educational purposes, which makes it an ideal resource for students and early career scientists. This computational process is accompanied by an accessible web-based tool with a graphical user interface that makes metabolomics data analysis accessible to non-experts and enables them to obtain statistical data- number on their data in a few minutes.
“This tool is accessible to a wide range of researchers, from complete beginners to experts, and is designed to be used in conjunction with the molecular networking software that my group is developing,” in said co-author Mingxun Wang, assistant professor of computer science. engineering at UCR. “For beginners, the guidance and code we provide make it easy to understand the common steps of processing and analyzing data. For experts, it accelerates reproducible data analysis, enabling them to share their statistical analysis work and results.”
Petras explained that the research paper is unique, serving as a major educational resource organized by a research group called the Virtual Multiomics Lab, or VMOL. With over 50 participating scientists from around the world, VMOL is a community-driven, open-access community. It aims to simplify and democratize the process of chemical analysis, making it accessible to researchers worldwide, regardless of background or resources.
“I am very proud to see how this project has become something influential, involving experts and students from all over the world,” said Abzer Pakkir Shah, a doctoral student in Petras’ group and first author of the paper. “By removing physical and economic barriers, VMOL provides training in computational mass spectrometry and data science and aims to initiate research projects as a new form of collaborative science.”
All tools developed by the team are free and publicly available. The development of the software began during the summer school of metabolomics that is not expected in 2022 at the University of Tübingen, where the team also introduced VMOL.
Petras expects that the protocol will be very useful for environmental researchers as well as scientists working in the biomedical field and researchers doing medical studies in microbiome science.
“The versatility of our protocol extends to different areas and sample types, including compound chemistry, doping testing, and the detection of contamination in food, pharmaceuticals, and other industrial products,” he said. said so.
Petras received his bachelor’s degree in biotechnology from the University of Applied Sciences Darmstadt and his doctorate in biochemistry from the Technical University Berlin. He did postdoctoral research at UC San Diego, where he focused on the development of large-scale environmental metabolomics methods. In 2021, he started the Functional Metabolomics Lab at the University of Tübingen. In January 2024 he joined UCR, where his lab focuses on the development and application of methods based on mass spectrometry to detect and monitor chemical exchange within communities. parasites.
Additional information:
Abzer K. Pakkir Shah et al, Statistical analysis of molecular-based network results from non-targeted metabolomics data, Laws of Nature (2024). DOI: 10.1038/s41596-024-01046-3
Offered by the University of California – Riverside
Excerpt: New data science tool dramatically speeds up molecular analysis of our environment (2024, September 20) retrieved on September 20, 2024 from https://phys.org/news/2024-09- science-tool-greatly-molecular-analysis.html
This document is subject to copyright. Except for any legitimate activity for the purpose of private study or research, no part may be reproduced without written permission. Content is provided for informational purposes only.
#data #science #tool #greatly #accelerating #molecular #analysis #environment